Title: Getting NJ assessment data into R: part 1 in a series.
Date: 2015-04-13
Category: education
Tags: NJ, assessment, NJASK, HSPA, data_management, tutorial
Slug: reading-nj-assess-data-1
Author: Andrew Martin
```{r, echo=FALSE}
#SET THIS TO TRUE WHEN READY TO PUBLISH
ready_to_ship = TRUE
library(knitr)
hook_plot <- knit_hooks$get('plot')
knit_hooks$set(plot=function(x, options) {
if (!is.null(options$pelican.publish) && options$pelican.publish) {
x <- paste0("{filename}", x)
}
hook_plot(x, options)
})
opts_chunk$set(dev='Cairo_svg')
opts_chunk$set(pelican.publish=ready_to_ship)
```
New Jersey has switched over to the PARCC assessment, but from roughly 2005-2014 New Jersey took the "New Jersey Assessment of Skills and Knowledge," or more simply "the NJASK."
This data is a bit of a pain to work with. There's a page for each [year](http://www.state.nj.us/education/schools/achievement/14/) of assessment data, and for each year there's a [page](http://www.state.nj.us/education/schools/achievement/14/njask8/) per grade level. But wait! It gets better! If you download the Excel spreadsheet for each grade level (helpfully titled `state_summary.xls` for _every_ year and grade!), you'll find **5** tabs per file, because there are simply too many demographic/subject subgroups for Excel 2003 to handle.
 I would like to say that I never go down this road, but looking at my downloads folder, I'm apparently on `state_summary(15).xls`.
Yuck.
# there must be a better way
Fortunately the state publishes [fixed width](http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt) versions of these files that you can work with. They're a little intimidating at first, but they're actually (relatively) easy to read into R.
## welcome to the Hadleyverse
We can use the `read_fwf()` function from [`hadley/readr`](https://github.com/hadley/readr) to quickly read fixed width files into R.
First let's load some libraries:
```{r libraries, message=FALSE, warning=FALSE}
library(readr)
library(dplyr)
library(sqldf)
library(reshape2)
library(magrittr)
```
We can pass one of those fixed-width data file to `read_fwf`, and it will try to convert it into an R data frame:
```{r fwf1}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_empty(file=sample_file)
)
as.data.frame(njask14_gr8)[1:11,1:15]
```
That's just the first fifteen of `r ncol(njask14_gr8)` columns -- we got some data into R (good), but those column names are awful unhelpful. Also, there's a bunch of weird `*` characters in our data. How can we identify each column?
# this part is unfortunate
For whatever reason NJ has seen fit to publish the crucial _definitions_ (column headers) for that data in an Excel spreadsheet. Never in my life have I seen something _more_ clearly suited for JSON.
I would like to say that I never go down this road, but looking at my downloads folder, I'm apparently on `state_summary(15).xls`.
Yuck.
# there must be a better way
Fortunately the state publishes [fixed width](http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt) versions of these files that you can work with. They're a little intimidating at first, but they're actually (relatively) easy to read into R.
## welcome to the Hadleyverse
We can use the `read_fwf()` function from [`hadley/readr`](https://github.com/hadley/readr) to quickly read fixed width files into R.
First let's load some libraries:
```{r libraries, message=FALSE, warning=FALSE}
library(readr)
library(dplyr)
library(sqldf)
library(reshape2)
library(magrittr)
```
We can pass one of those fixed-width data file to `read_fwf`, and it will try to convert it into an R data frame:
```{r fwf1}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_empty(file=sample_file)
)
as.data.frame(njask14_gr8)[1:11,1:15]
```
That's just the first fifteen of `r ncol(njask14_gr8)` columns -- we got some data into R (good), but those column names are awful unhelpful. Also, there's a bunch of weird `*` characters in our data. How can we identify each column?
# this part is unfortunate
For whatever reason NJ has seen fit to publish the crucial _definitions_ (column headers) for that data in an Excel spreadsheet. Never in my life have I seen something _more_ clearly suited for JSON.
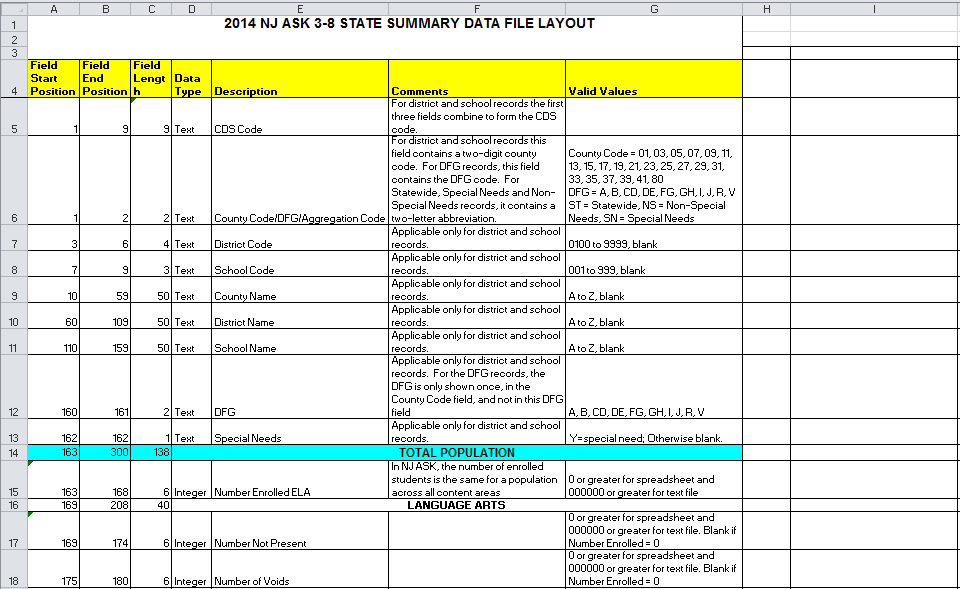 I think we can do some dplyr tricks and recover the nested aspects of this data. First, read the excel file in as a data frame:
```{r read_layout}
layout <- readr::read_csv("datasets/njask_layout.csv")
names(layout) <- tolower(gsub(' ', '_', names(layout)))
head(layout)
```
I did a little pre-processing in Excel before saving that csv -- namely, I created an indicator variable that indicates if a row was a 'spanning' row (basically, did the value span multiple columns?). The strategy here is going to be:
1) Separate those 'spanners' (which contain info, but aren't unique data elements) from the 'keepers' (the rows that uniquely ID each column).
2) Join our keepers back to our spanners, exploting the data about field start position and end position.
```{r munge1}
spanners <- dplyr::filter(layout, structural==TRUE)
keepers <- dplyr::filter(layout, structural==FALSE)
head(spanners)
```
To match spanners with keepers, we can do a SQL join (dplyr doesn't join on inequalities [yet](https://github.com/hadley/dplyr/issues/557)).
```{r}
with_spanners <- sqldf('
SELECT keepers.*
,spanners.data_type AS spanner
,spanners.field_length AS spanner_length
FROM keepers
LEFT OUTER JOIN spanners
ON keepers.field_start_position >= spanners.field_start_position
AND keepers.field_end_position <= spanners.field_end_position
')
with_spanners[1:20, c(1:5,8:10)]
```
So we have fields matched with their parents, but the sql join leaves us with _long_ data, and we really need _wide_ data (one row per field). Let's reshape!
First let's tag each row with a row number. For cases where we have multiple matching spanners(`TOTAL POPULATION` and `LANGUAGE ARTS`, for instance), this will let us collapse those rows into one record, with a column for `description`, `spanner1`, and `spanner2`.
```{r reshape1}
with_rn <- with_spanners %>%
dplyr::group_by(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values
) %>%
mutate(
rn = order(desc(spanner_length))
) %>%
select(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values, spanner, rn
) %>%
as.data.frame()
with_rn$rn <- paste0('spanner', with_rn$rn)
#mask NAS
with_rn$spanner <- ifelse(is.na(with_rn$spanner),'', with_rn$spanner)
head(with_rn)
```
We'll pass the row number we just generated as an identifier to one of the many functions in the `reshape2` [toolbox](http://www.cookbook-r.com/Manipulating_data/Converting_data_between_wide_and_long_format/#from-long-to-wide) (TBH I don't know any of those by heart - I just look up the syntax when needed).
```{r reshape2}
layout_njask <- dcast(
data = with_rn,
formula = field_start_position + field_end_position + field_length +
data_type + description + comments + valid_values ~ rn,
value.var = "spanner"
)
#this appears to be a bug in dcast? should not be needed.
layout_njask$spanner2 <- ifelse(is.na(layout_njask$spanner2),'', layout_njask$spanner2)
layout_njask[1:20, ]
```
Success! Now we just concatenate `description`, `spanner1`, and `spanner2` into a single value, do a little cleanup, and we have our headers. For cleanup, we want to get rid of weird whitespace, and convert reserved characters (like `+`) to words.
```{r combine}
#no plus or () symbols
layout_njask$spanner1 <- gsub('+', 'and', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub('(', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub(')', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$final_name <- layout_njask %$% paste(spanner1, spanner2, description, sep='_')
#kill double underscores
layout_njask$final_name <- gsub('__', '_', layout_njask$final_name)
#kill leading or trailer underscores
layout_njask$final_name <- gsub("(^_+|_+$)", "", layout_njask$final_name)
#trim any remaining whitespace
layout_njask$final_name <- gsub("^\\s+|\\s+$", "", layout_njask$final_name)
#all whitespace becomes underscore
layout_njask$final_name <- gsub(' ', '_', layout_njask$final_name)
#more whitespace cleanup
layout_njask$comments <- gsub("^\\s+|\\s+$", "", layout_njask$comments)
layout_njask$description <- gsub("^\\s+|\\s+$", "", layout_njask$description)
layout_njask$valid_values <- gsub("^\\s+|\\s+$", "", layout_njask$valid_values)
head(layout_njask)
```
Now we can hand our data frame of headers back to fwf using `fwf_positions` and get meaningful data:
```{r fwf2}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_positions(
start = layout_njask$field_start_position,
end = layout_njask$field_end_position,
col_names = layout_njask$final_name
),
na = "*"
)
```
success!
let's look at just a sample of 10 rows and 10 columns from that:
```{r fwf3}
dplyr::sample_n(njask14_gr8[, sample(c(1:551), 10)], 10) %>% as.data.frame()
```
In the [next post]({filename}/05_njask-data-2.Rmd), I'll talk about how to turn this linear script into a function that can process all of the state data. In preparation, let's dump the layout data frame that we cleaned up here to an `.rda` object so that we can easily use it later.
```{r export}
save(layout_njask, file = 'datasets/njask_layout.rda')
```
# appendix: all 551 headers
```{r all_the_headers}
layout_njask$final_name
```
I think we can do some dplyr tricks and recover the nested aspects of this data. First, read the excel file in as a data frame:
```{r read_layout}
layout <- readr::read_csv("datasets/njask_layout.csv")
names(layout) <- tolower(gsub(' ', '_', names(layout)))
head(layout)
```
I did a little pre-processing in Excel before saving that csv -- namely, I created an indicator variable that indicates if a row was a 'spanning' row (basically, did the value span multiple columns?). The strategy here is going to be:
1) Separate those 'spanners' (which contain info, but aren't unique data elements) from the 'keepers' (the rows that uniquely ID each column).
2) Join our keepers back to our spanners, exploting the data about field start position and end position.
```{r munge1}
spanners <- dplyr::filter(layout, structural==TRUE)
keepers <- dplyr::filter(layout, structural==FALSE)
head(spanners)
```
To match spanners with keepers, we can do a SQL join (dplyr doesn't join on inequalities [yet](https://github.com/hadley/dplyr/issues/557)).
```{r}
with_spanners <- sqldf('
SELECT keepers.*
,spanners.data_type AS spanner
,spanners.field_length AS spanner_length
FROM keepers
LEFT OUTER JOIN spanners
ON keepers.field_start_position >= spanners.field_start_position
AND keepers.field_end_position <= spanners.field_end_position
')
with_spanners[1:20, c(1:5,8:10)]
```
So we have fields matched with their parents, but the sql join leaves us with _long_ data, and we really need _wide_ data (one row per field). Let's reshape!
First let's tag each row with a row number. For cases where we have multiple matching spanners(`TOTAL POPULATION` and `LANGUAGE ARTS`, for instance), this will let us collapse those rows into one record, with a column for `description`, `spanner1`, and `spanner2`.
```{r reshape1}
with_rn <- with_spanners %>%
dplyr::group_by(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values
) %>%
mutate(
rn = order(desc(spanner_length))
) %>%
select(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values, spanner, rn
) %>%
as.data.frame()
with_rn$rn <- paste0('spanner', with_rn$rn)
#mask NAS
with_rn$spanner <- ifelse(is.na(with_rn$spanner),'', with_rn$spanner)
head(with_rn)
```
We'll pass the row number we just generated as an identifier to one of the many functions in the `reshape2` [toolbox](http://www.cookbook-r.com/Manipulating_data/Converting_data_between_wide_and_long_format/#from-long-to-wide) (TBH I don't know any of those by heart - I just look up the syntax when needed).
```{r reshape2}
layout_njask <- dcast(
data = with_rn,
formula = field_start_position + field_end_position + field_length +
data_type + description + comments + valid_values ~ rn,
value.var = "spanner"
)
#this appears to be a bug in dcast? should not be needed.
layout_njask$spanner2 <- ifelse(is.na(layout_njask$spanner2),'', layout_njask$spanner2)
layout_njask[1:20, ]
```
Success! Now we just concatenate `description`, `spanner1`, and `spanner2` into a single value, do a little cleanup, and we have our headers. For cleanup, we want to get rid of weird whitespace, and convert reserved characters (like `+`) to words.
```{r combine}
#no plus or () symbols
layout_njask$spanner1 <- gsub('+', 'and', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub('(', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub(')', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$final_name <- layout_njask %$% paste(spanner1, spanner2, description, sep='_')
#kill double underscores
layout_njask$final_name <- gsub('__', '_', layout_njask$final_name)
#kill leading or trailer underscores
layout_njask$final_name <- gsub("(^_+|_+$)", "", layout_njask$final_name)
#trim any remaining whitespace
layout_njask$final_name <- gsub("^\\s+|\\s+$", "", layout_njask$final_name)
#all whitespace becomes underscore
layout_njask$final_name <- gsub(' ', '_', layout_njask$final_name)
#more whitespace cleanup
layout_njask$comments <- gsub("^\\s+|\\s+$", "", layout_njask$comments)
layout_njask$description <- gsub("^\\s+|\\s+$", "", layout_njask$description)
layout_njask$valid_values <- gsub("^\\s+|\\s+$", "", layout_njask$valid_values)
head(layout_njask)
```
Now we can hand our data frame of headers back to fwf using `fwf_positions` and get meaningful data:
```{r fwf2}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_positions(
start = layout_njask$field_start_position,
end = layout_njask$field_end_position,
col_names = layout_njask$final_name
),
na = "*"
)
```
success!
let's look at just a sample of 10 rows and 10 columns from that:
```{r fwf3}
dplyr::sample_n(njask14_gr8[, sample(c(1:551), 10)], 10) %>% as.data.frame()
```
In the [next post]({filename}/05_njask-data-2.Rmd), I'll talk about how to turn this linear script into a function that can process all of the state data. In preparation, let's dump the layout data frame that we cleaned up here to an `.rda` object so that we can easily use it later.
```{r export}
save(layout_njask, file = 'datasets/njask_layout.rda')
```
# appendix: all 551 headers
```{r all_the_headers}
layout_njask$final_name
``` I would like to say that I never go down this road, but looking at my downloads folder, I'm apparently on `state_summary(15).xls`.
Yuck.
# there must be a better way
Fortunately the state publishes [fixed width](http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt) versions of these files that you can work with. They're a little intimidating at first, but they're actually (relatively) easy to read into R.
## welcome to the Hadleyverse
We can use the `read_fwf()` function from [`hadley/readr`](https://github.com/hadley/readr) to quickly read fixed width files into R.
First let's load some libraries:
```{r libraries, message=FALSE, warning=FALSE}
library(readr)
library(dplyr)
library(sqldf)
library(reshape2)
library(magrittr)
```
We can pass one of those fixed-width data file to `read_fwf`, and it will try to convert it into an R data frame:
```{r fwf1}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_empty(file=sample_file)
)
as.data.frame(njask14_gr8)[1:11,1:15]
```
That's just the first fifteen of `r ncol(njask14_gr8)` columns -- we got some data into R (good), but those column names are awful unhelpful. Also, there's a bunch of weird `*` characters in our data. How can we identify each column?
# this part is unfortunate
For whatever reason NJ has seen fit to publish the crucial _definitions_ (column headers) for that data in an Excel spreadsheet. Never in my life have I seen something _more_ clearly suited for JSON.
I would like to say that I never go down this road, but looking at my downloads folder, I'm apparently on `state_summary(15).xls`.
Yuck.
# there must be a better way
Fortunately the state publishes [fixed width](http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt) versions of these files that you can work with. They're a little intimidating at first, but they're actually (relatively) easy to read into R.
## welcome to the Hadleyverse
We can use the `read_fwf()` function from [`hadley/readr`](https://github.com/hadley/readr) to quickly read fixed width files into R.
First let's load some libraries:
```{r libraries, message=FALSE, warning=FALSE}
library(readr)
library(dplyr)
library(sqldf)
library(reshape2)
library(magrittr)
```
We can pass one of those fixed-width data file to `read_fwf`, and it will try to convert it into an R data frame:
```{r fwf1}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_empty(file=sample_file)
)
as.data.frame(njask14_gr8)[1:11,1:15]
```
That's just the first fifteen of `r ncol(njask14_gr8)` columns -- we got some data into R (good), but those column names are awful unhelpful. Also, there's a bunch of weird `*` characters in our data. How can we identify each column?
# this part is unfortunate
For whatever reason NJ has seen fit to publish the crucial _definitions_ (column headers) for that data in an Excel spreadsheet. Never in my life have I seen something _more_ clearly suited for JSON.
 I think we can do some dplyr tricks and recover the nested aspects of this data. First, read the excel file in as a data frame:
```{r read_layout}
layout <- readr::read_csv("datasets/njask_layout.csv")
names(layout) <- tolower(gsub(' ', '_', names(layout)))
head(layout)
```
I did a little pre-processing in Excel before saving that csv -- namely, I created an indicator variable that indicates if a row was a 'spanning' row (basically, did the value span multiple columns?). The strategy here is going to be:
1) Separate those 'spanners' (which contain info, but aren't unique data elements) from the 'keepers' (the rows that uniquely ID each column).
2) Join our keepers back to our spanners, exploting the data about field start position and end position.
```{r munge1}
spanners <- dplyr::filter(layout, structural==TRUE)
keepers <- dplyr::filter(layout, structural==FALSE)
head(spanners)
```
To match spanners with keepers, we can do a SQL join (dplyr doesn't join on inequalities [yet](https://github.com/hadley/dplyr/issues/557)).
```{r}
with_spanners <- sqldf('
SELECT keepers.*
,spanners.data_type AS spanner
,spanners.field_length AS spanner_length
FROM keepers
LEFT OUTER JOIN spanners
ON keepers.field_start_position >= spanners.field_start_position
AND keepers.field_end_position <= spanners.field_end_position
')
with_spanners[1:20, c(1:5,8:10)]
```
So we have fields matched with their parents, but the sql join leaves us with _long_ data, and we really need _wide_ data (one row per field). Let's reshape!
First let's tag each row with a row number. For cases where we have multiple matching spanners(`TOTAL POPULATION` and `LANGUAGE ARTS`, for instance), this will let us collapse those rows into one record, with a column for `description`, `spanner1`, and `spanner2`.
```{r reshape1}
with_rn <- with_spanners %>%
dplyr::group_by(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values
) %>%
mutate(
rn = order(desc(spanner_length))
) %>%
select(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values, spanner, rn
) %>%
as.data.frame()
with_rn$rn <- paste0('spanner', with_rn$rn)
#mask NAS
with_rn$spanner <- ifelse(is.na(with_rn$spanner),'', with_rn$spanner)
head(with_rn)
```
We'll pass the row number we just generated as an identifier to one of the many functions in the `reshape2` [toolbox](http://www.cookbook-r.com/Manipulating_data/Converting_data_between_wide_and_long_format/#from-long-to-wide) (TBH I don't know any of those by heart - I just look up the syntax when needed).
```{r reshape2}
layout_njask <- dcast(
data = with_rn,
formula = field_start_position + field_end_position + field_length +
data_type + description + comments + valid_values ~ rn,
value.var = "spanner"
)
#this appears to be a bug in dcast? should not be needed.
layout_njask$spanner2 <- ifelse(is.na(layout_njask$spanner2),'', layout_njask$spanner2)
layout_njask[1:20, ]
```
Success! Now we just concatenate `description`, `spanner1`, and `spanner2` into a single value, do a little cleanup, and we have our headers. For cleanup, we want to get rid of weird whitespace, and convert reserved characters (like `+`) to words.
```{r combine}
#no plus or () symbols
layout_njask$spanner1 <- gsub('+', 'and', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub('(', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub(')', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$final_name <- layout_njask %$% paste(spanner1, spanner2, description, sep='_')
#kill double underscores
layout_njask$final_name <- gsub('__', '_', layout_njask$final_name)
#kill leading or trailer underscores
layout_njask$final_name <- gsub("(^_+|_+$)", "", layout_njask$final_name)
#trim any remaining whitespace
layout_njask$final_name <- gsub("^\\s+|\\s+$", "", layout_njask$final_name)
#all whitespace becomes underscore
layout_njask$final_name <- gsub(' ', '_', layout_njask$final_name)
#more whitespace cleanup
layout_njask$comments <- gsub("^\\s+|\\s+$", "", layout_njask$comments)
layout_njask$description <- gsub("^\\s+|\\s+$", "", layout_njask$description)
layout_njask$valid_values <- gsub("^\\s+|\\s+$", "", layout_njask$valid_values)
head(layout_njask)
```
Now we can hand our data frame of headers back to fwf using `fwf_positions` and get meaningful data:
```{r fwf2}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_positions(
start = layout_njask$field_start_position,
end = layout_njask$field_end_position,
col_names = layout_njask$final_name
),
na = "*"
)
```
success!
let's look at just a sample of 10 rows and 10 columns from that:
```{r fwf3}
dplyr::sample_n(njask14_gr8[, sample(c(1:551), 10)], 10) %>% as.data.frame()
```
In the [next post]({filename}/05_njask-data-2.Rmd), I'll talk about how to turn this linear script into a function that can process all of the state data. In preparation, let's dump the layout data frame that we cleaned up here to an `.rda` object so that we can easily use it later.
```{r export}
save(layout_njask, file = 'datasets/njask_layout.rda')
```
# appendix: all 551 headers
```{r all_the_headers}
layout_njask$final_name
```
I think we can do some dplyr tricks and recover the nested aspects of this data. First, read the excel file in as a data frame:
```{r read_layout}
layout <- readr::read_csv("datasets/njask_layout.csv")
names(layout) <- tolower(gsub(' ', '_', names(layout)))
head(layout)
```
I did a little pre-processing in Excel before saving that csv -- namely, I created an indicator variable that indicates if a row was a 'spanning' row (basically, did the value span multiple columns?). The strategy here is going to be:
1) Separate those 'spanners' (which contain info, but aren't unique data elements) from the 'keepers' (the rows that uniquely ID each column).
2) Join our keepers back to our spanners, exploting the data about field start position and end position.
```{r munge1}
spanners <- dplyr::filter(layout, structural==TRUE)
keepers <- dplyr::filter(layout, structural==FALSE)
head(spanners)
```
To match spanners with keepers, we can do a SQL join (dplyr doesn't join on inequalities [yet](https://github.com/hadley/dplyr/issues/557)).
```{r}
with_spanners <- sqldf('
SELECT keepers.*
,spanners.data_type AS spanner
,spanners.field_length AS spanner_length
FROM keepers
LEFT OUTER JOIN spanners
ON keepers.field_start_position >= spanners.field_start_position
AND keepers.field_end_position <= spanners.field_end_position
')
with_spanners[1:20, c(1:5,8:10)]
```
So we have fields matched with their parents, but the sql join leaves us with _long_ data, and we really need _wide_ data (one row per field). Let's reshape!
First let's tag each row with a row number. For cases where we have multiple matching spanners(`TOTAL POPULATION` and `LANGUAGE ARTS`, for instance), this will let us collapse those rows into one record, with a column for `description`, `spanner1`, and `spanner2`.
```{r reshape1}
with_rn <- with_spanners %>%
dplyr::group_by(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values
) %>%
mutate(
rn = order(desc(spanner_length))
) %>%
select(
field_start_position, field_end_position, field_length,
data_type, description, comments, valid_values, spanner, rn
) %>%
as.data.frame()
with_rn$rn <- paste0('spanner', with_rn$rn)
#mask NAS
with_rn$spanner <- ifelse(is.na(with_rn$spanner),'', with_rn$spanner)
head(with_rn)
```
We'll pass the row number we just generated as an identifier to one of the many functions in the `reshape2` [toolbox](http://www.cookbook-r.com/Manipulating_data/Converting_data_between_wide_and_long_format/#from-long-to-wide) (TBH I don't know any of those by heart - I just look up the syntax when needed).
```{r reshape2}
layout_njask <- dcast(
data = with_rn,
formula = field_start_position + field_end_position + field_length +
data_type + description + comments + valid_values ~ rn,
value.var = "spanner"
)
#this appears to be a bug in dcast? should not be needed.
layout_njask$spanner2 <- ifelse(is.na(layout_njask$spanner2),'', layout_njask$spanner2)
layout_njask[1:20, ]
```
Success! Now we just concatenate `description`, `spanner1`, and `spanner2` into a single value, do a little cleanup, and we have our headers. For cleanup, we want to get rid of weird whitespace, and convert reserved characters (like `+`) to words.
```{r combine}
#no plus or () symbols
layout_njask$spanner1 <- gsub('+', 'and', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub('(', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$spanner1 <- gsub(')', '', layout_njask$spanner1, fixed = TRUE)
layout_njask$final_name <- layout_njask %$% paste(spanner1, spanner2, description, sep='_')
#kill double underscores
layout_njask$final_name <- gsub('__', '_', layout_njask$final_name)
#kill leading or trailer underscores
layout_njask$final_name <- gsub("(^_+|_+$)", "", layout_njask$final_name)
#trim any remaining whitespace
layout_njask$final_name <- gsub("^\\s+|\\s+$", "", layout_njask$final_name)
#all whitespace becomes underscore
layout_njask$final_name <- gsub(' ', '_', layout_njask$final_name)
#more whitespace cleanup
layout_njask$comments <- gsub("^\\s+|\\s+$", "", layout_njask$comments)
layout_njask$description <- gsub("^\\s+|\\s+$", "", layout_njask$description)
layout_njask$valid_values <- gsub("^\\s+|\\s+$", "", layout_njask$valid_values)
head(layout_njask)
```
Now we can hand our data frame of headers back to fwf using `fwf_positions` and get meaningful data:
```{r fwf2}
sample_file = "http://www.state.nj.us/education/schools/achievement/14/njask8/state_summary.txt"
njask14_gr8 <- readr::read_fwf(
file = sample_file,
col_positions = readr::fwf_positions(
start = layout_njask$field_start_position,
end = layout_njask$field_end_position,
col_names = layout_njask$final_name
),
na = "*"
)
```
success!
let's look at just a sample of 10 rows and 10 columns from that:
```{r fwf3}
dplyr::sample_n(njask14_gr8[, sample(c(1:551), 10)], 10) %>% as.data.frame()
```
In the [next post]({filename}/05_njask-data-2.Rmd), I'll talk about how to turn this linear script into a function that can process all of the state data. In preparation, let's dump the layout data frame that we cleaned up here to an `.rda` object so that we can easily use it later.
```{r export}
save(layout_njask, file = 'datasets/njask_layout.rda')
```
# appendix: all 551 headers
```{r all_the_headers}
layout_njask$final_name
```